Diagnostics
Intra- and inter-chain diagnostics can tell us how well a particular inference algorithm performed on the model. Two common diagnostics are effective sample size, and R-hat.1
R-hat
is a diagnostic tool that measures the between- and within-chain variances. It is a test that indicates a lack of convergence by comparing the variance between multiple chains to the variance within each chain. If the parameters are successfully exploring the full space for each chain, then , since the between-chain and within-chain variance should be equal. is calculated from samples as
where is the within-chain variance, is the between-chain variance and is the estimate of the posterior variance of the samples. The take-away here is that converges to 1 when each of the chains begins to empirically approximate the same posterior distribution. We do not recommend using inference results if . More information about can be found in the reference 2.
Effective Sample Size (ESS)
MCMC samplers do not draw truly independent samples from the target distribution, which means that our samples are correlated. In an ideal situation all samples would be independent, but we do not have that luxury. We can, however, measure the number of effectively independent samples we draw, which is called the effective sample size. You can read more about how this value is calculated in the [2] paper. In brief, it is a measure that combines information from the value with the autocorrelation estimates within the chains.
ESS estimates come in two variants, ess_bulk and ess_tail. The former is
the default, but the latter can be useful if you need good estimates of the
tails of your posterior distribution. The rule of thumb for ess_bulk is for
this value to be greater than 100 per chain on average. Since we ran four
chains, we need ess_bulk to be greater than 400 for each parameter. The
ess_tail is an estimate for effectively independent samples considering the
more extreme values of the posterior. This is not the number of samples that
landed in the tails of the posterior, but rather a measure of the number of
effectively independent samples if we sampled the tails of the posterior. The
rule of thumb for this value is also to be greater than 100 per chain on
average.
Diagnostics information with ArviZ
We can use ArviZ, a third-party package for exploratory analysis of Bayesian models, to provide helpful statistics about the result of the inference algorithm, including R-hat and ESS.
To do that, we wrap the result of Bean Machine inference into an InferenceData ArviZ object and use its methods for diagnostic information:
posterior = bm.SingleSiteNewtonianMonteCarlo().infer(
queries,
observations,
num_samples,
num_chains,
)
import arviz as az
inference_data = az.convert_to_inference_data(posterior.samples)
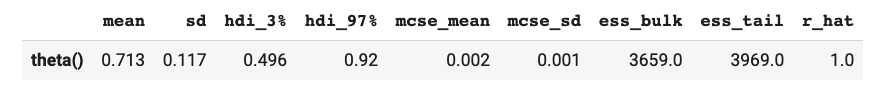
az.summary(inference_data)
which in a notebook outputs something like:
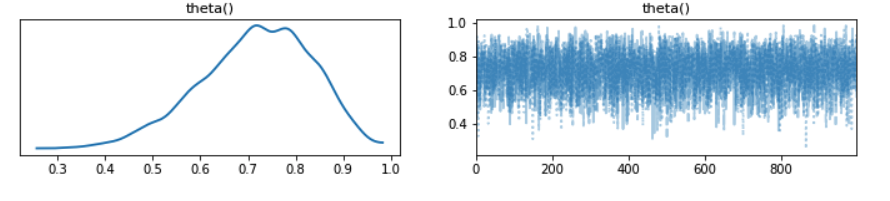
ArviZ provides many other analysis tools, including trace and autocorrelation plots:
az.plot_trace(inference_data);
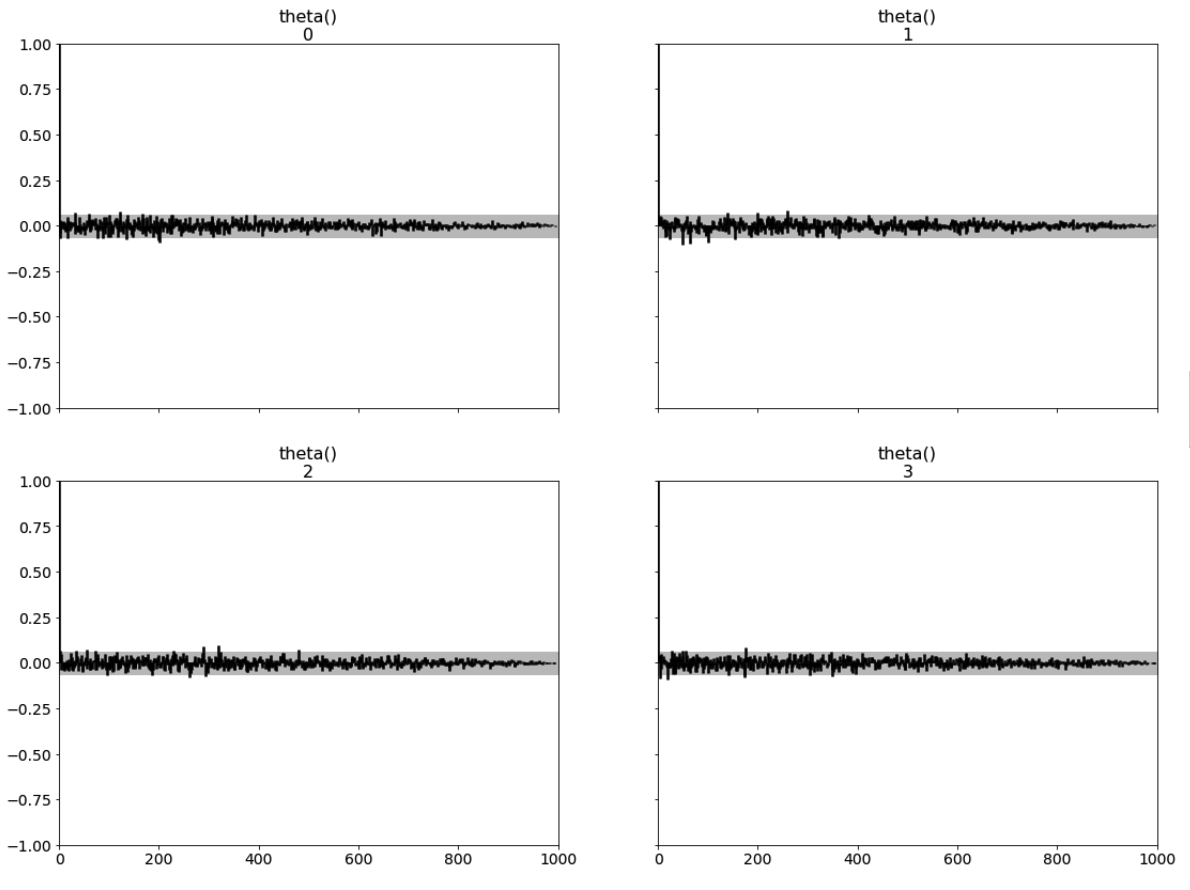
az.plot_autocorr(inference_data, combined=False, max_lag=1000, grid=(2,2));
1 Stan Reference Manual. https://mc-stan.org/docs/2_18/reference-manual/effective-sample-size-section.html
2 Vehtari A, Gelman A, Simpson D, Carpenter B, Bürkner PC (2021) Rank-Normalization, Folding, and Localization: An Improved for Assessing Convergence of MCMC (with Discussion). Bayesian Analysis 16(2) 667–718. doi: 10.1214/20-BA1221.